When Stanislav Dusko Ehrlich – a world expert in microbiology and a pioneer of metagenomics – and his team set out to create their next generation biotech research platform, they needed a technology solution to support their stringent capacity and performance requirements for big data analytics. This use case provides insight into the requirements and decision making process for applying big data technology toward genomic research.
National Institute of Agronomic Research
National Institute of Agronomic Research (INRA) in France coordinates and hosts MetaGenoPolis (MGP), a unique facility in Europe at the service of the medical, scientific and industrial communities. Under the direction of Ehrlich, the MGP team has established recognized scientific landmarks. Their research paves the way for early diagnosis and new therapies for many diseases such as diabetes, obesity and metabolic diseases generally associated with our digestive system. They study a still little known “organ”: the intestinal microbiota, the billions of bacteria that inhabit our digestive tract.
Genome Analytics
A key aspect of MGP’s success is that it is designed as a fully integrated project, from the sequencing materials and methods through to the data pre-processing and analytic algorithms. Therefore in addition to MGP’s performance and scalability requirements were added the constraints of full autonomy and self-sufficiency from a technical management point of view.
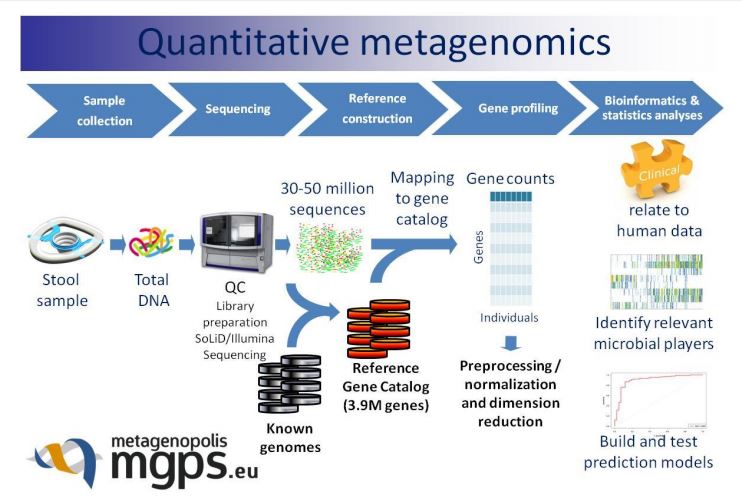
The platform’s data processing layout is similar to that of many labs :
- Raw data pipeline: preliminary processing of raw data, in this case from DNA sequencers in a pipeline designed to associate, aggregate and annotate the data against reference sets on a record level (e.g. clinical information) as well as on a data set level (e.g. clustering).
- Interactive data analytics: data exploration and research by teams of biologists and bio-statisticians.
These two phases have very different performance requirements. The pipeline requires ultra-high throughput/data import rates. Interactive data analytics on the other requires the capacity to filter, select and perform sub-second aggregations on data within a peta-scale data store. The technology chosen to address these requirements involved solutions from ParStream.
During benchmarks, ParStream boosted performance by a factor of 100x across both throughput and responsiveness” says Jean-Michel Batto, MGP’s IT lead. “The goal of MGP is not only to be able to scale by a factor of 10 the individual DNA sample volumes but furthermore to intersect the cohorts from which the samples are draws. A file based solution could neither support the scale nor the functionality, with ParStream we will be able to investigate biological data in much greater depth and breadth.”
Increased pipeline throughput will allow MGP to increase the scope of their research by a factor of 10x while accelerated analytics will enable cross cohort research directly on fine grain data.
The ParStream real time database – able to run on any configuration from bare-iron to virtualized cloud instances – not only allows users to select commodity hardware according to their target usage but furthermore allows more flexibility that any other alternative by supporting CPU-GPU computing. Along with the simplicity of a SQL interface, MGP’s research team is thus totally autonomous and self-sufficient in the management and evolution of their stack, integrating it as their project and activities demand. The possibility of serving INRA MGP’s peta-scale requirements with commodity hardware while achieving with the combination of HPCI and GPU the highest levels of energy efficiency were essential ParStream differentiators.
At the heart of ParStream performance is an innovation in Big Data indexing. ParStream’s HPCI (High Performance Compressed Index) has significant properties :
- Indexes are compressed and never decompressed, this reduces memory and bandwidth requirements as well CPU cycle requirements by avoiding decompression.
- Indexes processing is multi-core/multi-server/multi-site, this increases the responsiveness to queries as well as providing the flexibility of a federated database perspective for end-users.
- Indexes are bit-vector based thus allowing researchers total flexibility in the expression of their queries without requiring specific system tuning and in fact response time that accelerates with query complexity and selectiveness.
Further to the performance requirements, hardware flexibility expectations and resource efficiency, INRA MGP demands the possibility of implementing their own in-database algorithms in order to support highly computational and data intense operations such as clustering without overloading standard cluster interconnects. ParStream highly optimized cluster communication and UDF options through a C++ API set rounded out the functionality that INRA MGP required.
INRA MGP wanted much more than a packaged DBMS. Packaged DBMSs are good for the category of applications who’s structure, content and meaning are already well understood. A research team on the other hand progresses in a non-linear manner and seeks to find patterns and relationships in an apparently amorphous collection of data and information.
Sign up for the free insideBIGDATA newsletter.





Speak Your Mind